This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
Model organisms having homologous CDH23 proteins
Cow [Bos taurus]
Acccession: XM_001252944
FASTA
Chicken [Gallus gallus]
Acccession: XP_421595
FASTA
Chimpanzee [Pan troglodytes]
Acccession: XP_507839
FASTA
Acccession: XM_001252944
FASTA
Chicken [Gallus gallus]
Acccession: XP_421595
FASTA
Chimpanzee [Pan troglodytes]
Acccession: XP_507839
FASTA
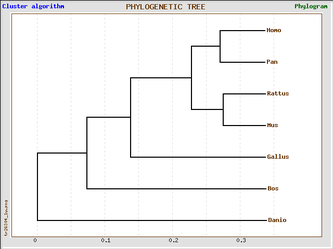
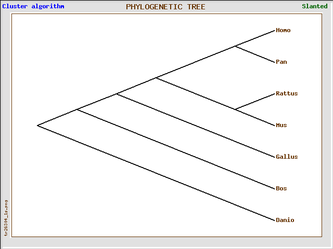
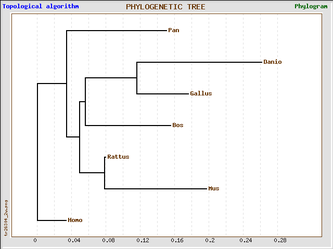
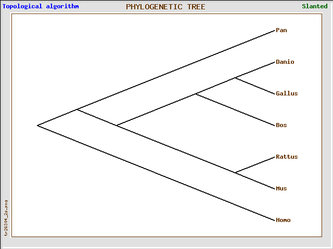
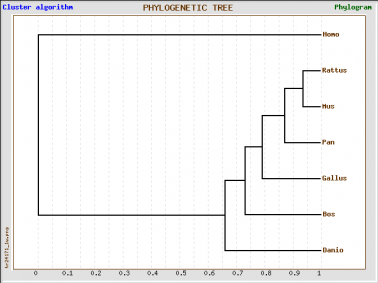
Phylogenic trees of Homo sapien and six model organisms based on cadherin-like 23 (Click on image to enlarge)
The 4 phylogenic trees are all produced from the same data set using GeneBee. A clustered algorithim was used for the first two and a topological algorithm for the last two. Trees with perpendicular and parallel lines are phylograms, and diagonal trees are slanted trees.
Analysis:
Clustered algorithm trees tell us that, in the evolutionary process, Pan troglodyte is our most closely related ancestor, because we share the most recent last common ancestor. We are also in closer relation with the rat and mouse, who themselves share the most recent last common ancestor. These trees also indicate that Danio rerio (the zebrafish) is the most distant relative of all six model organisms.
Topological trees show mostly the same information, with the exception of Homo sapien's close relationship with Pan troglodyte. These trees show immediate divergence of the human with other species.
Topological algorithm trees take connectivity into consideration first, before branch length is concerned, whereas cluster algorithm trees first take into account distance between groups.
Analysis:
Clustered algorithm trees tell us that, in the evolutionary process, Pan troglodyte is our most closely related ancestor, because we share the most recent last common ancestor. We are also in closer relation with the rat and mouse, who themselves share the most recent last common ancestor. These trees also indicate that Danio rerio (the zebrafish) is the most distant relative of all six model organisms.
Topological trees show mostly the same information, with the exception of Homo sapien's close relationship with Pan troglodyte. These trees show immediate divergence of the human with other species.
Topological algorithm trees take connectivity into consideration first, before branch length is concerned, whereas cluster algorithm trees first take into account distance between groups.
Alignments of Human CDH23 and other Model Organisms From Entrez
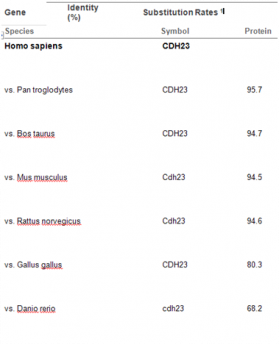
Alignment percentages from BLASTing sequences in Entrez support the data provided by phyolgenic trees. The alignment shows greater similarity to Bos taurus that the mouse and rat. This discrepancy is likely do to the difference in algorithms implemented on each site.
Comparing finalized CDH23 with cadherin-like 23 proteins of model organisms
Using GeneBee once again I ran a phylogenic comparison for the six model organisms against the finalized CDH23 Homo sapien protein. The tree shows a large divergence of the human from the rest of the organisms, indicating that between the human and the 6 model organisms, completely different evolutionary paths were taken that produced proteins of similar function. This pattern is seen in our topological algorithm trees, however a different set of relationships exist for the six model organisms. I will neglect this data for my study.
References:
NCBI Protein Databasae: http://www.ncbi.nlm.nih.gov/sites/entrez?db=Protein&itool=toolbar
Entrez: http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=homologene&dopt=AlignmentScores&list_uids=11142
NCBI Protein Databasae: http://www.ncbi.nlm.nih.gov/sites/entrez?db=Protein&itool=toolbar
Entrez: http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=homologene&dopt=AlignmentScores&list_uids=11142
Ben Hofeld, [email protected], last updated: 5.15.2010, Link to course page:www.gen677.weebly.com