This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
Experiment Overview
My future experiment will be studying wild-type and mutant cdh23 proteins, in an attempt to asses the domain requirements for normal protein function. Specifically, I will be analyzing the mus muluscus cadherin 23 precursor protein located in inner ear hair cells. Besides the Rhesus Monkey, the mouse is the model organism that possesses the greatest homology to human cdh23 precursor protein. In studying these proteins, I will be performing qualitative mass spectrometry to determine full-length sequences for both wild-type and mutant groups. With a respectable sample size I will align all sequences and, using bioinformatic software, determine the likelihood of certain domains, domain locations, or domain abundance, being requirements for a functional final cdh23 protein.
Experimental subjects, generating sample size, and testing function

The Jackson Lab in Bar Harbor, Maine is a major distributor of laboratory mice. Currently, two cdh23 mutant mice which express type 1 Usher Syndrome are available for purchase, and are hghlighted below:
----------------------------------------------------------------------------------------------------------------------------------------
JAX® Mice models Adato et al. (2005) wrote an excellent review of the five mutant genes known to be responsible for Usher syndrome. We distribute JAX® Mice models for each:
B6.A-Ush1g js/J ("Jackson Shaker," 000783) for sans
SH1/LeJ ("shaker," 000271) for myosin VIIa,
B6;129S4-Ush1cdfcr-2J/J and BALB/cBy-Ush1cdfcr/J ("deaf circler," 004768 and 004771) for harmonin
B6J x B6.C-H2bm1/ByJ-Cdh23v-J/J andC57BL/6J-Cdh23v-2J/J ("waltzer," 002432 and 002552 respectively) for cadherin 23, and
C57BL/6J-Pcdh15av-3J/J ("Ames waltzer," 002072) for protocadherin 15
-----------------------------------------------------------------------------------------------------------------------------------------
Protein sequences are not available for these mutants, so our assessment of the sequence could be novel in itself.Further, after the two strains are sequenced, we will need to build our sample size for a meaningful comparison. I will take some of each of the mutant strains and induce further mutations. The type of mutation/deletion f I will likely be making is dependent on the preliminary proteomics data, but mutations that effectively insert lost (from mutation) cadherin domains, that are conserved in both mice and humans, seems to be a logical target. Inducing a mutation in a mutant that increases its homology to the wild-type could cause a regain of function.
I will take the opposite approach for wild-type mice. After preliminary proteomics on WT mice I will induce mutations resulting in deletion of protein domains or point mutations of inter-membrane protein sections to determine if a specific domain knock-out or residue alteration results in a loss of function. Normal wild-type mice are also available for purchase at Jackson Lab. (http://jaxmice.jax.org/research/neurobiology/usher.html)
Loss of function will be diagnosed by no response to a tuning fork being sounded close to the ear. The control group will help account for deafness caused by other means, if the issue arises.
*Sample size can be expanded if such protein sequences from Jax mice are reported in primary literature.*
----------------------------------------------------------------------------------------------------------------------------------------
JAX® Mice models Adato et al. (2005) wrote an excellent review of the five mutant genes known to be responsible for Usher syndrome. We distribute JAX® Mice models for each:
B6.A-Ush1g js/J ("Jackson Shaker," 000783) for sans
SH1/LeJ ("shaker," 000271) for myosin VIIa,
B6;129S4-Ush1cdfcr-2J/J and BALB/cBy-Ush1cdfcr/J ("deaf circler," 004768 and 004771) for harmonin
B6J x B6.C-H2bm1/ByJ-Cdh23v-J/J andC57BL/6J-Cdh23v-2J/J ("waltzer," 002432 and 002552 respectively) for cadherin 23, and
C57BL/6J-Pcdh15av-3J/J ("Ames waltzer," 002072) for protocadherin 15
-----------------------------------------------------------------------------------------------------------------------------------------
Protein sequences are not available for these mutants, so our assessment of the sequence could be novel in itself.Further, after the two strains are sequenced, we will need to build our sample size for a meaningful comparison. I will take some of each of the mutant strains and induce further mutations. The type of mutation/deletion f I will likely be making is dependent on the preliminary proteomics data, but mutations that effectively insert lost (from mutation) cadherin domains, that are conserved in both mice and humans, seems to be a logical target. Inducing a mutation in a mutant that increases its homology to the wild-type could cause a regain of function.
I will take the opposite approach for wild-type mice. After preliminary proteomics on WT mice I will induce mutations resulting in deletion of protein domains or point mutations of inter-membrane protein sections to determine if a specific domain knock-out or residue alteration results in a loss of function. Normal wild-type mice are also available for purchase at Jackson Lab. (http://jaxmice.jax.org/research/neurobiology/usher.html)
Loss of function will be diagnosed by no response to a tuning fork being sounded close to the ear. The control group will help account for deafness caused by other means, if the issue arises.
*Sample size can be expanded if such protein sequences from Jax mice are reported in primary literature.*
Discussion of potential results
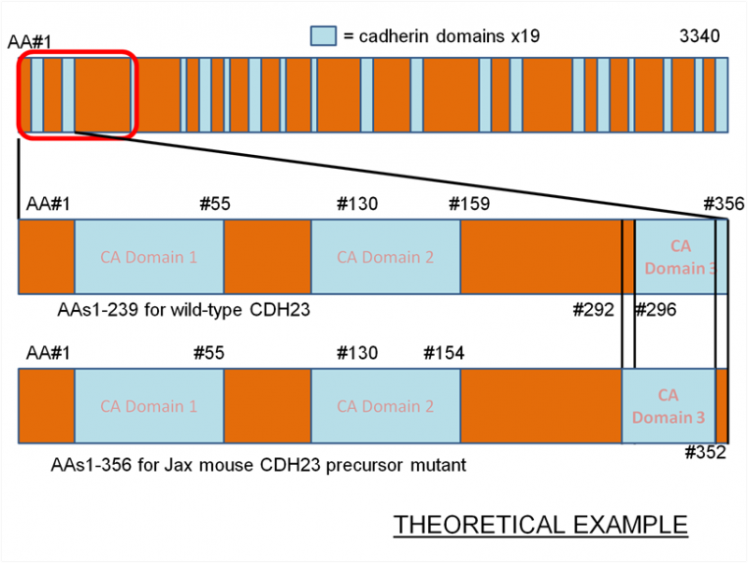
As an example, imagine that in the diagram below, the THEORETICAL 3340 AA protein represents the wild-type cdh23 precursor for Jax mice. The second band represents a blown-up version of the first 356 AAs of this wildtype protein precursor. The third band represents the first 356 AAs of a mutant aligned with that of the wildtype. Given that the only difference between the two proteins is the loss of amino acids 292-296 from WT to mutant, we can suggest something about something about the cause for loss of functionality.
We can suggest that:
1) The relative distance in residues between CA domains 1 and 2 (or a further domain) affect the functionality of the protein
OR
2) The loss of amino acids 154-158 in THEMSELVES caused the loss of function
1) The relative distance in residues between CA domains 1 and 2 (or a further domain) affect the functionality of the protein
OR
2) The loss of amino acids 154-158 in THEMSELVES caused the loss of function
Other Ideas and Future Directions
Human CDH23 in audio hair cells grown from stem cells
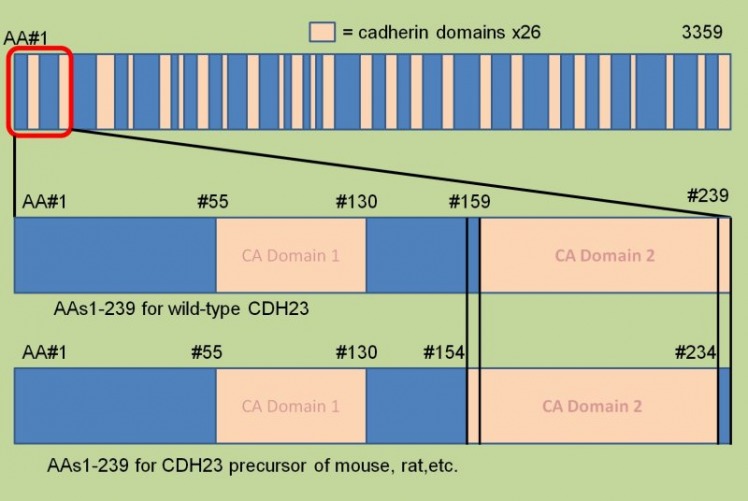
This is an ACTUAL EXAMPLE in which the homologies between human cdh23 precursor protein and that of the mouse and rat only differ in the deletions of amino acids 144-148 and the point mutation of AA 149 from V-->N. The resulting alignment shift is seen above. Because the rest of the precursor protein is conserved among a handful of model organisms, this region could be an evelutionary adaptation, and it would be interesting to test if the WT preservation of 144-149 is a requirement for normal function. (NCBI Homologene)
Binding sites of CDH23, USH1C,
MYO7A, and USH2A
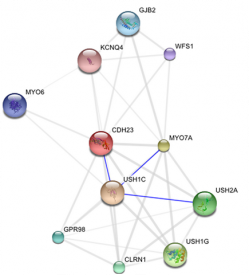
STRING 8.2
If you recall from the protein network interactions tab, the primary and secondary interaction proteins of CDH23 are all related through protein-protein binding, though the protein regions for these bindings are largely unknown. Understanding the binding sites for each protein binding interaction would give us clues as to which parts of the proteins are most important for proper function. To start researching this we would have to look at:
- protein foldings, structure of CDH23 and USH1C
- Role of some proteins as chaperones
References:
Jackson Lab. (http://jaxmice.jax.org/research/neurobiology/usher.html)
NCBI Homologene: http://www.ncbi.nlm.nih.gov/sites/entrez?Db=homologene&DbFrom=gene&Cmd=Link&LinkName=gene_homologene&LinkReadableName=HomoloGene&IdsFromResult=64072
STRING 8.2: http://string-db.org/newstring_cgi/show_input_page.pl?UserId=122XyLCvXqZ_&sessionId=XyVnU_PF2Un7
Jackson Lab. (http://jaxmice.jax.org/research/neurobiology/usher.html)
NCBI Homologene: http://www.ncbi.nlm.nih.gov/sites/entrez?Db=homologene&DbFrom=gene&Cmd=Link&LinkName=gene_homologene&LinkReadableName=HomoloGene&IdsFromResult=64072
STRING 8.2: http://string-db.org/newstring_cgi/show_input_page.pl?UserId=122XyLCvXqZ_&sessionId=XyVnU_PF2Un7
Ben Hofeld, [email protected], last updated: 5.15.2010, Link to course page:www.gen677.weebly.com